Research
My research is broadly in the area of statistical genetics (list of publications). Specific topics of interest include:
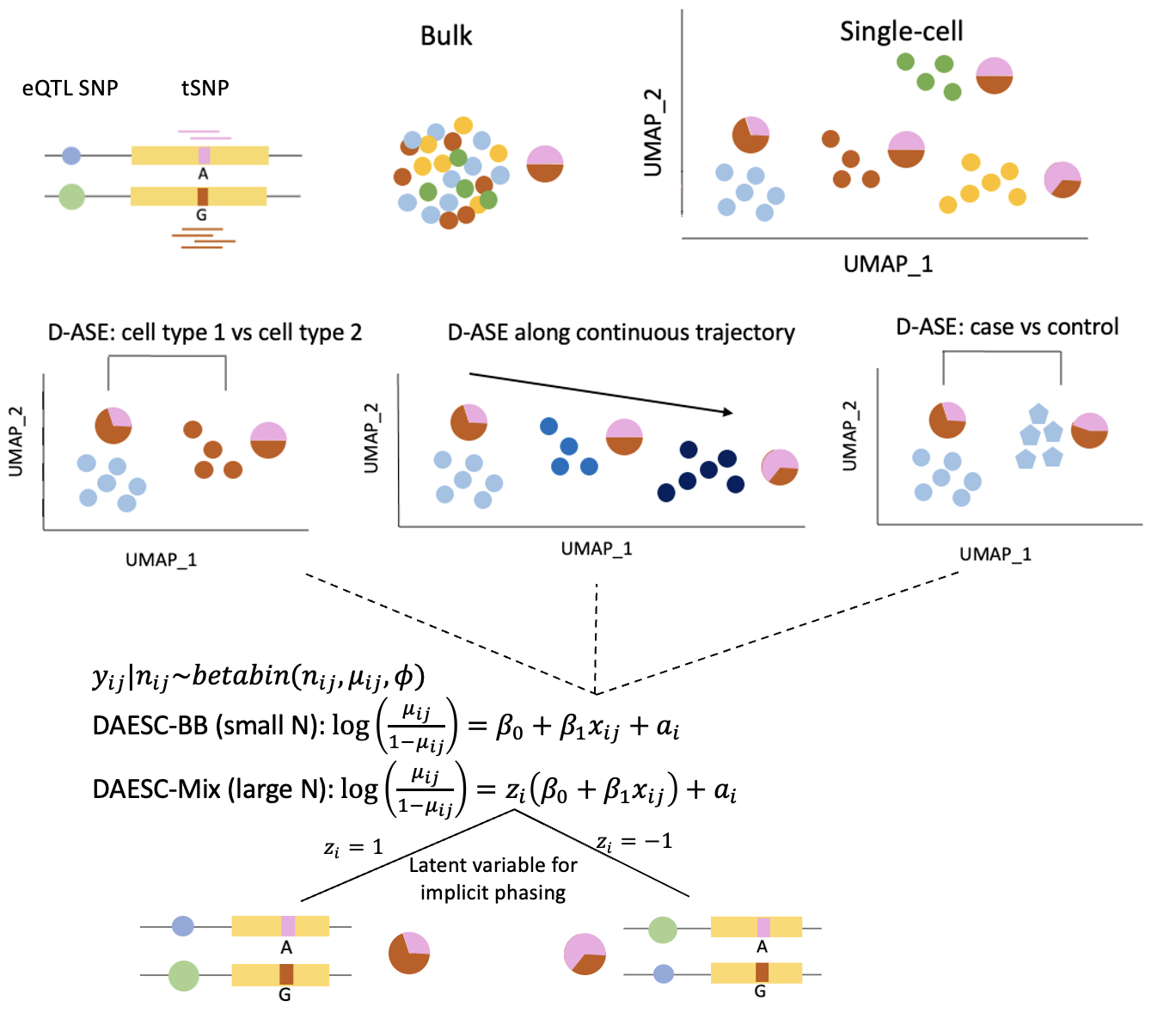
Integration of single-cell eQTL and GWAS data
GWAS have identified thousands of variants associated with complex traits and diseases. However, the underlying biological mechanisms are unclear. Single-cell expression quantitative trait loci (eQTL) and allele-specific expression (ASE) data characterize the effect of genetic variants on gene expression at cell-type-specific level, and hence reveal potential mediating mechanisms for variant-trait associations. Statistical methods typically involve high-dimensional and functional data analysis.
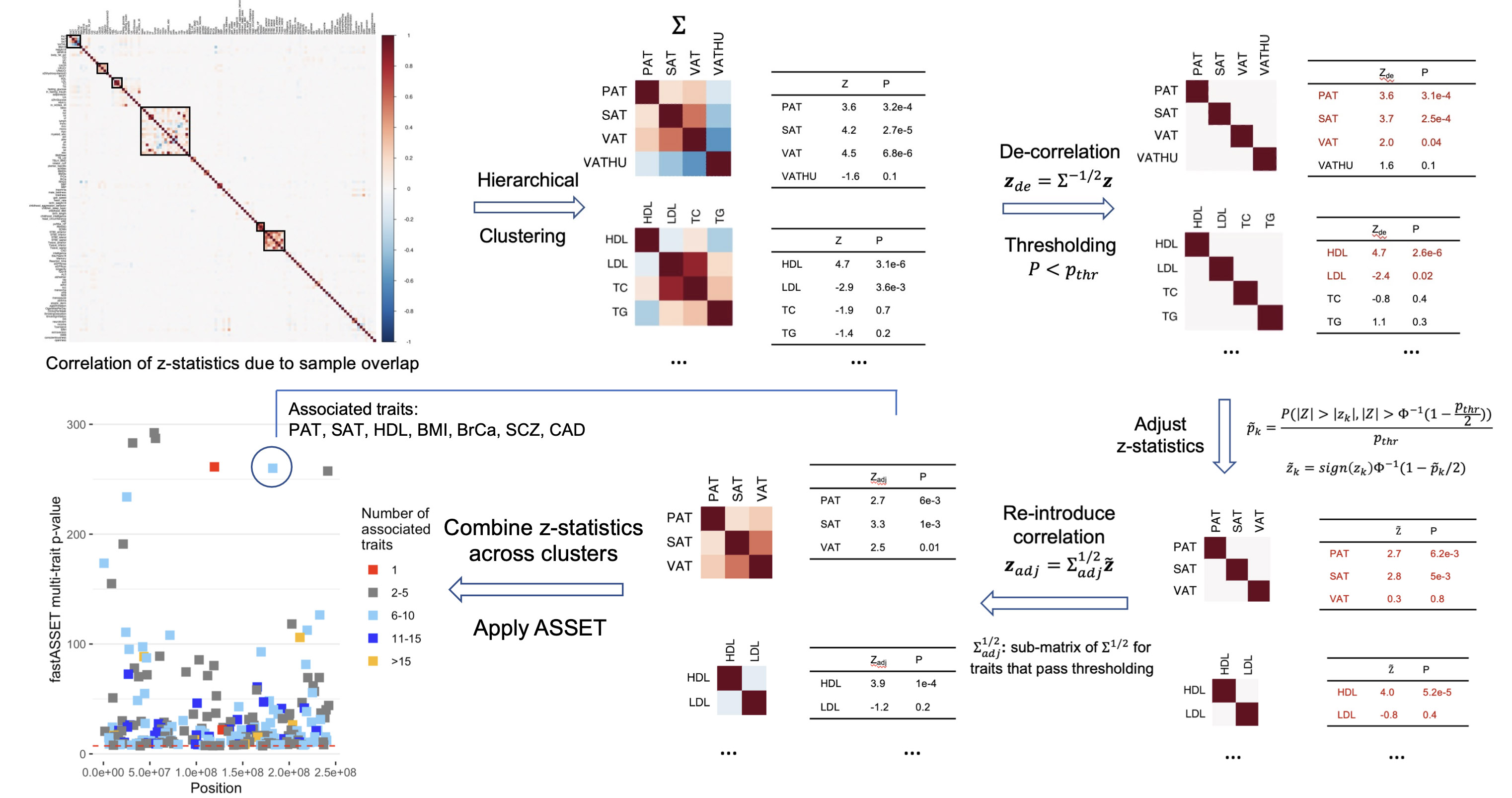
Multi-trait genetic association analysis
Traditionally, GWAS analyze one trait at time to investigate its genetic architecture. Multi-trait methods integrate information across traits to yield novel insight of variant function across the phenome and boost statistical power. Statistical methods for this area involve empirical Bayes methods, meta-analysis, etc.
Mendelian randomization
Mendelian randomization (MR), which lies in the intersection of statistical genetics and causal inference, uses genetic variants as instrumental variables to study the causal relationship between risk factors and diseases. My work includes MR methods robust to horizontal pleiotropy and multi-ethnic MR.